Table of contents
4) Discussion
4.1 Urban waste water treatment and AMR
The major known sources and transmission routes for AMR are through health and food applications. Pathways to the environment through urban waste water treatment largely arise from people excreting resistant bacteria themselves, or taking medicine and excreting some of the active ingredient, which may allow bacteria in the environment to develop resistance.
Gene transfer can take place in urban waste water treatment plants, though the rate and extent may be dependent on a range of factors, including the type of treatment applied. The longer the effluent is stored, the more opportunity there may be for AMR to be generated. This creates challenges: for instance, while disinfection is used to reduce the bacterial load, the waiting time between secondary treatment and disinfection can allow resistant bacteria to grow.
Disinfection can affect different genes in different ways (Fig [C]). Particularly in the context of urban waste water, it should be noted that the presence of other substances, such as some metals, can co-select for resistance [EDITING NOTE - why does this matter? super-resistant BUGS OR SOMETHING ELSE?].
Fig [C] Number of antibiotic resistant genes following disinfection treatment of urban waste water effluents – ozonation and UV.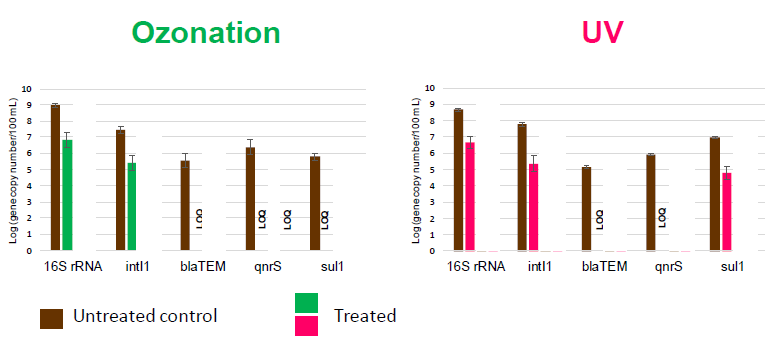
Source: Sousa et al, J. Hazard Mater. 2017 5;323(Pt A):434-441.
LOQ = below limit of quantification.
Selected genes 16S rRNA gene (marker for bacteria), the intl 1 gene (maker for class 1 integrons, common in Gram-negative bacteria), the blaTEM gene (marker for beta-lactamase resistant Gram-negative bacteria) and the antibiotic resistant genes qnrS and sul1.
A study in the Netherlands considered 100 UWWTPs (a third of all Dutch UWWTPs) and loadings in the context of well-characterised understanding of water across the country[1]. UWWT reduced the number of bacteria 100-1000 times and was significantly correlated with E. coli. Most of the differences in removal efficiency can be explained by rainfall, where increased rainfall can flush more waste into sewers but can also dilute the influent.
Some work suggests that urban waste water from hospitals represents a potential hotspot for AMR. However, the study showed that usually hospital effluents were not a very significant proportion of influent concentrations (below 10%), although some showed antibiotic concentrations over the minimum inhibitory concentration.
In a study considering treated effluents from UWWTPs in six countries (China, Estonia, Finland, Portugal, Spain and USA), Manaia et al. (2016) found that the numbers of antibiotic resistant bacteria and antibiotic resistant genes released each day varied between plants with a range of 6 orders of magnitude (1012-1018)[2]. While this is still a large number, significant reduction of the loads released was considered an adequate strategy to mitigate antibiotic resistance.
4.2 Pathways and exposure in the environment
There is limited information on the pathways for and significance of AMR in the environment to reach humans. Because genes can be so readily transferred between microorganisms, there may be several stages involved in any pathway.
A reservoir of AMR may exist in microorganisms that rarely reach humans, but these may interact with “carriers” which readily acquire and transfer genes. These carriers can interact with “vectors” which are widespread, human associated bacteria which can pass on genes to pathogens which have high capacity to infect and cause disease. The physical routes back into humans or animals, then, are through ingestion of contaminated (raw) food and drinking water; physical contact between people/animals, or from surfaces in public areas, recreation (water, sand) or health care. The remaining way is through inhalation, where sanitation workers would be most at risk of inhaling droplets, or populations downwind of intensive farms which have been shown to be more likely to be colonised by animal associated AMR bacteria. Fig [D] provides examples of these different pathways.
Fig [D] Possible routes of transmission of AMR back into humans.
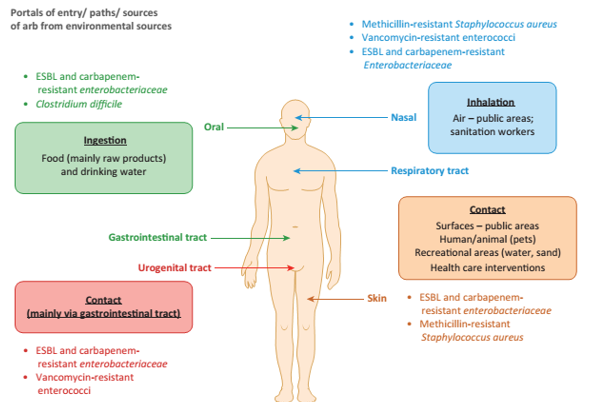
Source: Manaia, Trends Microbiol. 2017 25(3):173-181.
A key knowledge gap is the significance of environmental exposure of AMR to humans, relative to health and food pathways. However this issue is complicated by the fact that relatively rare gene transfer events from environmental bacteria may lead to pandemic spread of AMR in human associated bacteria. The threshold for environmental levels becoming of concern in terms of acute exposure risk needs to be assessed, although studies such as that by Leonard et al., (2018)[3] suggest that environmental exposure and transmission does occur. Better understanding of the drivers for resistance propagation and persistence is required, and we need to identify which resistance determinants should be of most concern.
Escherichia coli (E. coli) are widely used as indicators of fecal pollution of water. Extended spectrum beta-lactase (ESBL)-producing E. coli are representative of enzymes produced by bacteria to destroy antibiotics and are one way that bacteria develop resistance. Certain groups of people have greater exposure to such resistant bacteria, principally those swimming in fresh or coastal waters e.g. surfers[4].
4.3 Monitoring
Robust monitoring procedures are key to ensuring the reliability and comparability of results between different workers and sites. The relative novelty of AMR in the environment means broadly applicable monitoring methods and strategies applicable have yet to be agreed.
Clarity of the monitoring purpose is essential. For AMR in the environment, there are different possibilities. To protect human and domestic animal health, monitoring could assess prevalence of the level of resistance, the use of antibiotics, or the transmission or evolution of resistance, for example. For environmental protection, some measure of ecological effect would be more appropriate. Selection of suitable sites follows the purpose. Monitoring programmes also need to consider costs, method robustness and skills required to implement the methods.
[1] rivm.nl/bibliotheek/rapporten/2017-0058
[2] Manaia et al., 2016. Applied Microbiology and Biotechnology. 100:1543–1557
[3] https://doi.org/10.1016/j.envint.2017.11.003
[4] Leonard et al, 2017. DOI: 10.1016/j.envint.2015.02.013